|
|
DNA Extraction
M 1 2 3 4 5 6 7 8 9 10 11
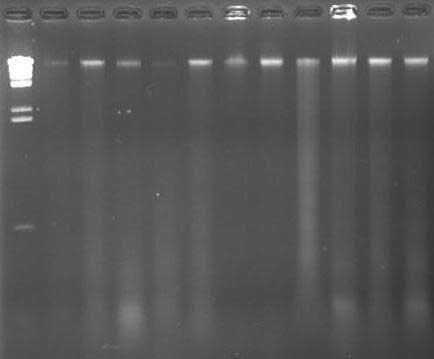 M: l Hind III DNA Marker; Lanes 1 ミ 11: DNA Extract
M: l Hind III DNA Marker; Lanes 1 ミ 11: DNA Extract
( 0.8% Agarose Gel: TBE: 75 Volts: 60 Mins )
DNA was extracted using a modified protocol based on Dellaporta
|
5ユ- Anchored PCR Amplification
1 2 M 3 4
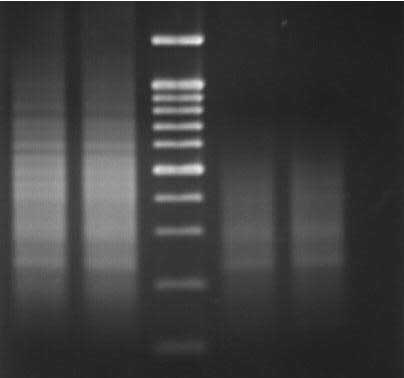 200bp
M: 100 Bp. Marker; Lanes 1,2: Mg++ (2.5 mM); Lanes
3, 4: Mg++ (1.5 mM) 200bp
M: 100 Bp. Marker; Lanes 1,2: Mg++ (2.5 mM); Lanes
3, 4: Mg++ (1.5 mM)
( 1.2% Agarose Gel: TBE: 75 Volts: 60 Mins )
5ユ Anchored PCR amplification can be used to successfully
amplify regions of DNA which contain microsatellite Loci.
|
Plasmid RE Digestion
M 1 2 3 4 5 6 7 M 8 9 10 11
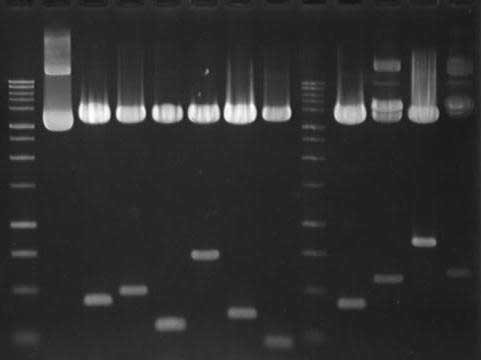 M: 100 bp DNA Ladder; Lanes 1 - 11: Plamid s digested
with EcoRI (Promega)
M: 100 bp DNA Ladder; Lanes 1 - 11: Plamid s digested
with EcoRI (Promega)
( 1.0 % Agarose Gel: TBE: 75 Volts: 60 Mins )
|
Microsatellite Locus
 Microsatellite locus showing 5ユ and 3ユ terminal
repeats.
Microsatellite locus showing 5ユ and 3ユ terminal
repeats.
|
Microsatellite Loci
Clone 5ユ Terminal Repeat Internal repeat(s) 3ユ
Terminal Repeat
|
Clone |
5' Terminal Repeat |
Internal repeat(s) |
3' Terminal Repeat |
|
PRA04 |
(AC) 10 |
(AT) 7 |
(GT) 13 |
|
PRA06 |
(AC) 11 |
-- |
-- |
|
PRA08 |
(AC) 10 |
(GT) 4 |
( GT) 11 |
|
PRA09 |
(AC) 11 |
(GT) 6 |
(GT) 10 |
|
PRA11 |
(AC) 10 |
-- |
-- |
|
PRA12 |
(AC)10 |
(AC) 4 |
(GT) 11 |
|
PRA17 |
(AC) 11 |
-- |
-- |
|
PRC03 |
(AC) 10 |
(GA) 8 |
(GT) 10 |
|
PRC09 |
(AC) 11 |
-- |
(GT) 11 |
|
PRC10 |
(AC) 11 |
-- |
(GT) 13 |
|
PRC11 |
(AC) 10 |
(AT) 5 |
(GT) 10 |
|
PRC12 |
(AC)12 |
(GT) 6 |
(GT) 10 |
|
PRC13 |
(AC) 10 |
-- |
-- |
|
PRC14 |
(AC) 12 |
-- |
(GT) 10 |
|
PRC15 |
(AC) 10 |
(CT) 7 |
(GT) 10 |
|
PRC 16 |
(AC) 10 |
-- |
(GT) 10 |
|
PRC18 |
(AC) 12 |
-- |
(GT) 10 |
|
PRC 19 |
(AC) 13 |
(AT) 5 |
(GT) 10 |
|
PRC 20 |
(AC) 10 |
-- |
(GT) 11 |
43 Microsatellite Loci were characterized for the first time
in Paphiopedilum rothschildianum, these comprise 34
terminal repeat sequences located at the 5ユ and 3ユ end and 9
internal repeat sequences.
|
|
|
| The 5ユ-Anchored PCR Method has proven to be
an efficient, rapid and cost-effective method in isolating and
characterizing microsatellite loci in genomes where no prior
DNA sequence data is available. A total of 43 microsatellite
loci were isolated and characterized. |
REFERENCES
1. Dellaporta S. L., Wood J., Hicks, J. B., 1983. A Plant
DNA Minipreparation: Version II. Plant Molecular Biology Reporter
1: 19 - 21.
2. Fisher, P. J. , Gardener, R.C., Richardson, T.E., 1996. Single
locus microsatellites isolated using 5ユ Anchored PCR. Nucleic
Acid Research. 24: 4369 - 4371
3. Kumar, S. V., Tan, S. G.,Quah, S. C., Yusoff, K. 2002. Isolation
and characterization of seven tetranucleotide microsatellite
loci in mungbean, Vigna radiata. Molecular Ecology Notes
2: 293 -295.
4. Sambrook, J., Fritsch, E. F. and Maniatis, T. 1989. Molecular
Cloning: A Laboratory Manual, 2nd Edition, Cold Spring Harbor
Laboratory Press, CSH, NY. |
This project is funded by the Malaysian Government
IRPA Grant No. 01-02-10-0054-EA0052
The authors are grateful to the Sabah Parks Authority
for providing specimens of Paphiopedilum rothschildianum
from their in-situ conservation facilities at Poring and
Kinabalu. |
|
|
|
|

 200bp
200bp
